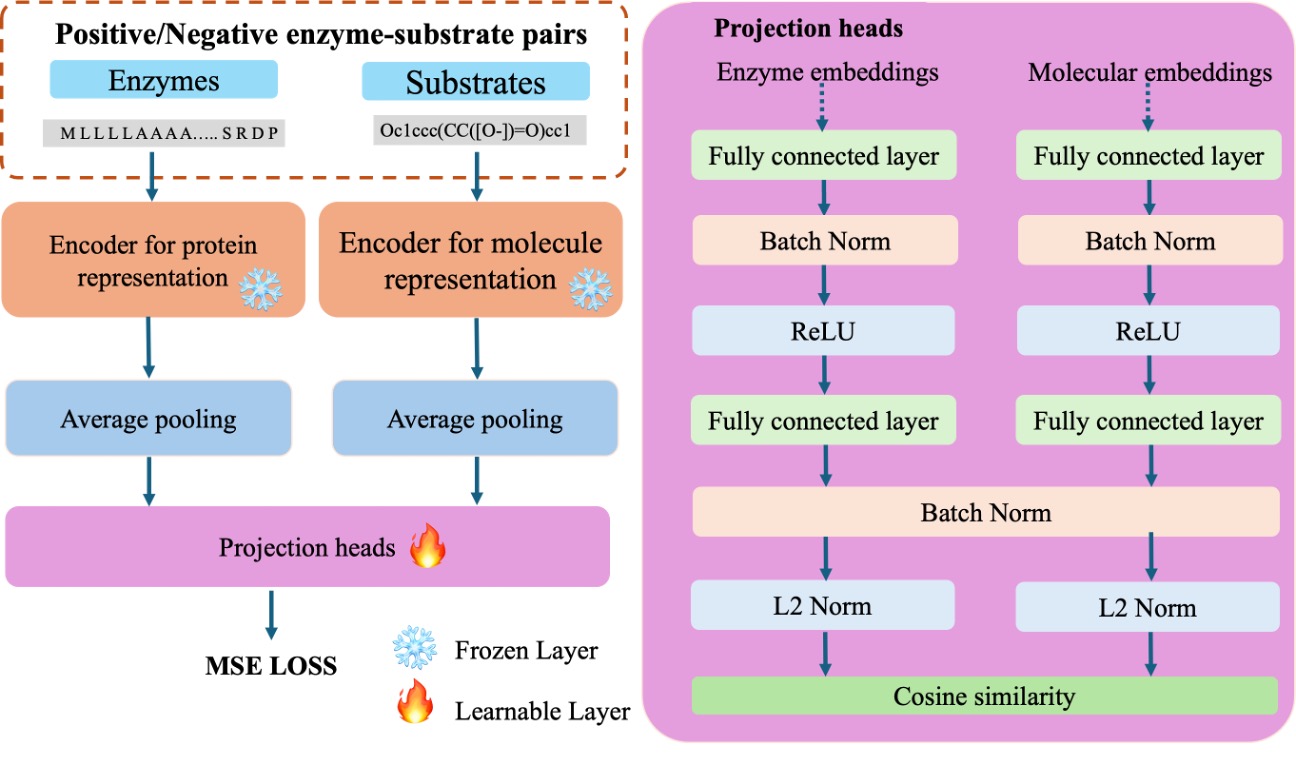
FusionESP: Improved enzyme-substrate pair prediction by fusing protein and chemical knowledge
The webserver is the implementation of the paper "FusionESP: Improved enzyme-substrate pair prediction by fusing protein and chemical knowledge". The original codes for the model development are available at https://github.com/dzjxzyd/FusionESP
Acknowledgment: Portions of this work were supported by the National Science Foundation CICI Program (2419880)
Notice: a confidence score will be given for you, which reflect how confidence the model think its output. The model deployed in this web server is the FusionESP-XL with ESM-2-1280 and MolFormer trained on both phylogenetic evidence-based and experimental evidence-based datasets (ACC = 94.56% AUC = 0.9635 MCC = 0.8572)
Large-scale output version: 1. Prepare your files (xls, xlsx) and click “Choose File” for uploading → 2. Choose a model for classification → 3. Download the results.
Usage of the webserver:
Example for “Quick output version” :
1. Insert a enzyme protein sequence (in single amino acid code), and a SMILES representation of molecules (canonical SMILES) → → → 2. Click “Run”→ → → 4. The result will be returned in seconds below the “Run” button
Notice: it also support multiple sequence at the same time. Just input as “VPP,IPP,CCL,AGR” (sequences are separated by comma, no space)
Example for “Large-scale output version:” :
1. Prepare your xls, xlsx, txt or fasta files → → → 2. Upload the file through “Choose File” botton → → → 3. Click “Run” → → → 4. It will automatically download your results.
Notice: File preparation should follow the examples under this repository https://github.com/dzjxzyd/FusionESP_server_1280/tree/main/input